Follow us on Google News (click on ☆)
These new findings pave the way for better management of this cancer and the development of more targeted treatments.

Glioblastoma is the most common but also the most aggressive brain tumor. Approximately 3,500 new cases are diagnosed each year in France. Despite many scientific advances, this tumor remains incurable due to its high molecular and cellular heterogeneity, which complicates the use of standard therapeutic approaches.
"The problem is that each tumor is unique: the genes expressed are numerous and different, forming a complex network of interactions. Our work reveals a hierarchy controlled by 'master regulators'—highly interconnected key molecules—that actively maintain the tumor," explains Mohamed Elati, head of the "Digital Systems & Cancer Computational" team at the CANTHER laboratory in Lille.
In an effort to categorize tumors to refine treatments, scientists had previously identified four tumor subgroups based on patients' transcriptional profiles (gene expression). However, some groups remained highly heterogeneous.
In this new study, researchers focused on the activity of regulatory molecules, transcription factors, which interact with our genes, activating or inhibiting their expression. Out of the 2,375 transcription factors and cofactors present in humans, 539 are active in glioblastoma tumor mechanisms.
Thanks to AI and machine learning in particular, researchers were able to unify data from 16 international studies conducted over several years (involving approximately 1,600 patients). This approach led to the creation of the largest mapping to date of glioblastoma transcriptional activity, identifying this time no fewer than seven tumor subtypes, each associated with specific biological mechanisms and different prognoses (see figure).
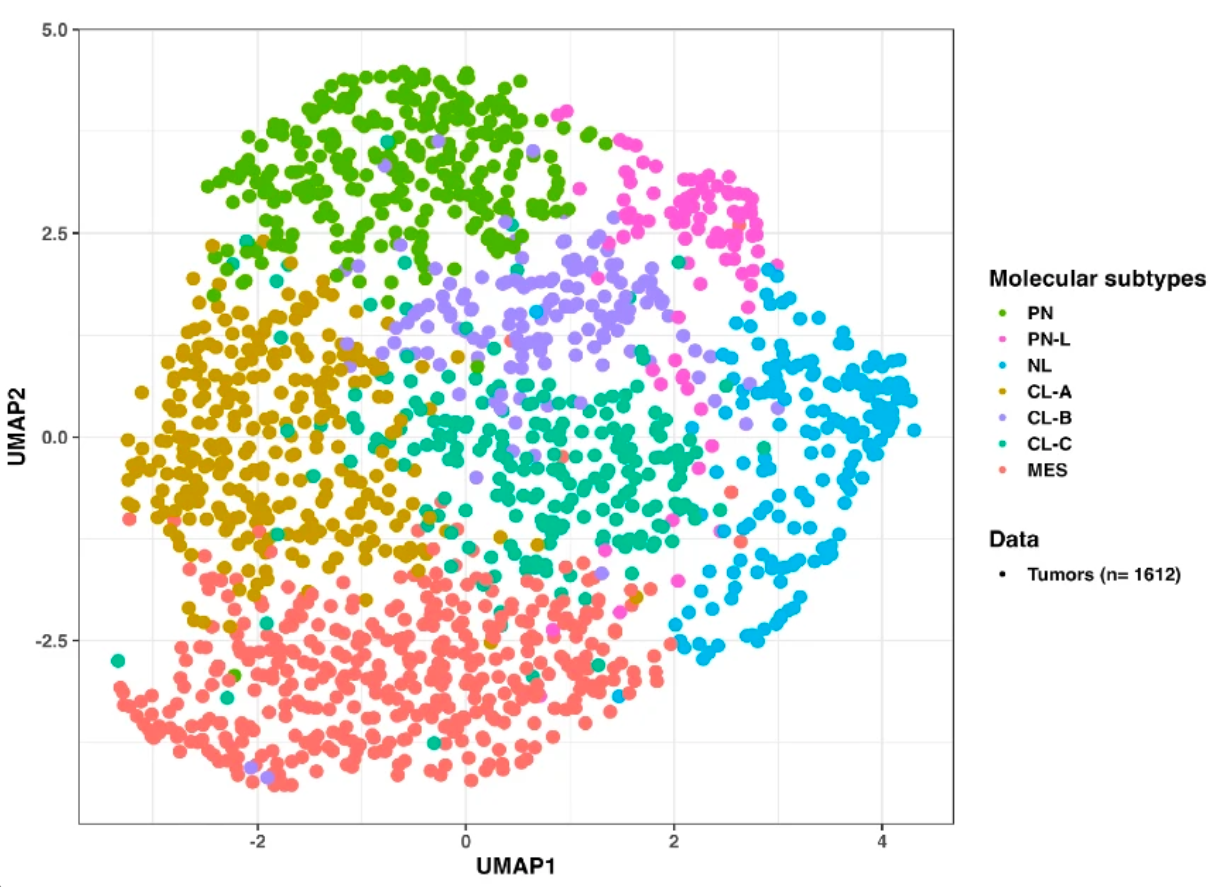
Mapping of transcription factor activity in the 1,600 recorded cases. Each color is associated with a specific tumor type and its unique mechanisms.
This bioinformatics tool, made available to the scientific community and named GBM-cRegMap, aims to precisely determine, based on individual molecular data, the characteristics of the tumor at the time of detection as well as after treatment during recurrence. These valuable insights will help better understand glioblastoma mechanisms and its progression, as well as the development of more personalized therapies.
This map also reveals that current preclinical models (cellular models simulating the tumor and allowing for the testing of new therapies) may not, in reality, cover all the identified tumor types, highlighting the need to develop new cell lines.