Follow us on Google News (click on ☆)
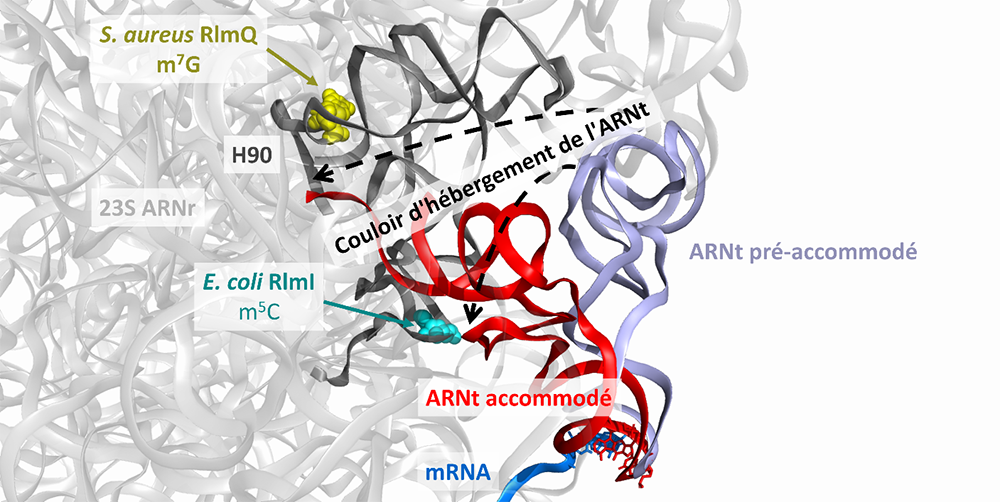
Figure: Divergent rRNA modifications (m7G2601 in Staphylococcus aureus, m5C1962 in Escherichia coli) in the tRNA accommodation corridor are governed by the enzymes RlmQ (S. aureus) and RlmI (E. coli), thus revealing translation subtleties unique to each species.
© Stefano Marzi
In an article published in the journal RNA, scientists unveil the existence of a new enzyme conserved in several Gram-positive bacteria (named for their positive reaction to Gram staining due to their wall characteristics). This enzyme, a methylase, specifically modifies a single nucleotide of the ribosomal RNA (rRNA) by adding a methyl group. This modification, which occurs during ribosome biogenesis, impacts the complex molecular machinery orchestrating protein synthesis. Positioned in the ribosome's corridor through which transfer RNAs (tRNAs) pass to provide the essential amino acids for protein construction, this modification occupies a strategic position.
A multidisciplinary approach including the use of artificial intelligence
This new modification enzyme was discovered through a multidisciplinary approach including genetic analyses, the use of artificial intelligence, and meticulous verification of RNA modifications. Surprisingly, this discovery revealed that the enzyme has a significant impact on specific virulence traits in golden staph or Staphylococcus aureus (S. aureus), a major human pathogen, highlighting a complex interaction between rRNA modifications, the translation process, and the mechanisms of virulence factor synthesis.
New avenues for the development of targeted therapies against bacterial infections
This study sheds light on the evolution of the roles of specific rRNA modification enzymes, which extend to adaptive responses during stress situations, environmental changes, and infections caused by pathogenic bacteria, thus forming an intriguing area ready to be explored. These discoveries emphasize the need to continue exploring the complex links between RNA modifications, the cellular machinery, and the responses of pathogenic bacteria to the various conditions they face. A better understanding of these mechanisms could open new avenues for the development of targeted therapies against bacterial infections, thus offering exciting perspectives for future research in the field of microbiology.
Reference:
Bahena-Ceron R, Teixeira C, Ponce JRJ, Wolff P, Couzon F, François P, Klaholz BP, Vandenesch F, Romby P, Moreau K, Marzi S. RlmQ: a newly discovered rRNA modification enzyme bridging RNA modification and virulence traits in Staphylococcus aureus. RNA. 2024 Feb 16;30(3):200-212. doi: 10.1261/rna.079850.123.