Follow us on Google News (click on ☆)
This method, called "epi-bits," promises unmatched storage density and increased efficiency. Researchers from Peking University recently published their work in Nature, paving the way for practical and personalized applications.

The team, led by Cheng Zhang and Long Qian, developed a technique to encode data as epigenetic modifications on DNA strands. This approach, named "epi-bits," uses enzymatic methylation to mark specific positions on universal DNA templates. Unlike traditional methods, this technique does not require de novo DNA synthesis (assembling the molecule's components one by one), making the process faster and less expensive.
One of the major advantages of this technology is its ability to store a phenomenal amount of information in a tiny space. A single gram of DNA can hold up to 215,000 terabytes of data, equivalent to 10 million hours of high-definition video. This storage density, combined with the long-term stability of DNA, makes it an ideal medium for data archiving.
The epi-bits method relies on the parallel assembly of pre-synthesized DNA fragments, called DNA bricks, onto a reusable strand. Each brick binds to a unique location, guiding an enzyme to methylate a specific position. This process allows data to be encoded in a binary system, similar to that used in computing.
The researchers demonstrated the efficiency of their method by encoding 275,000 bits of information on five DNA templates, without requiring long and costly DNA synthesis. Among the stored data were two high-definition photos, illustrating the potential of this technology for image and video storage. Additionally, a platform called iDNAdrive allowed volunteers to encode data themselves, with a read error rate of "only" 1.42%.
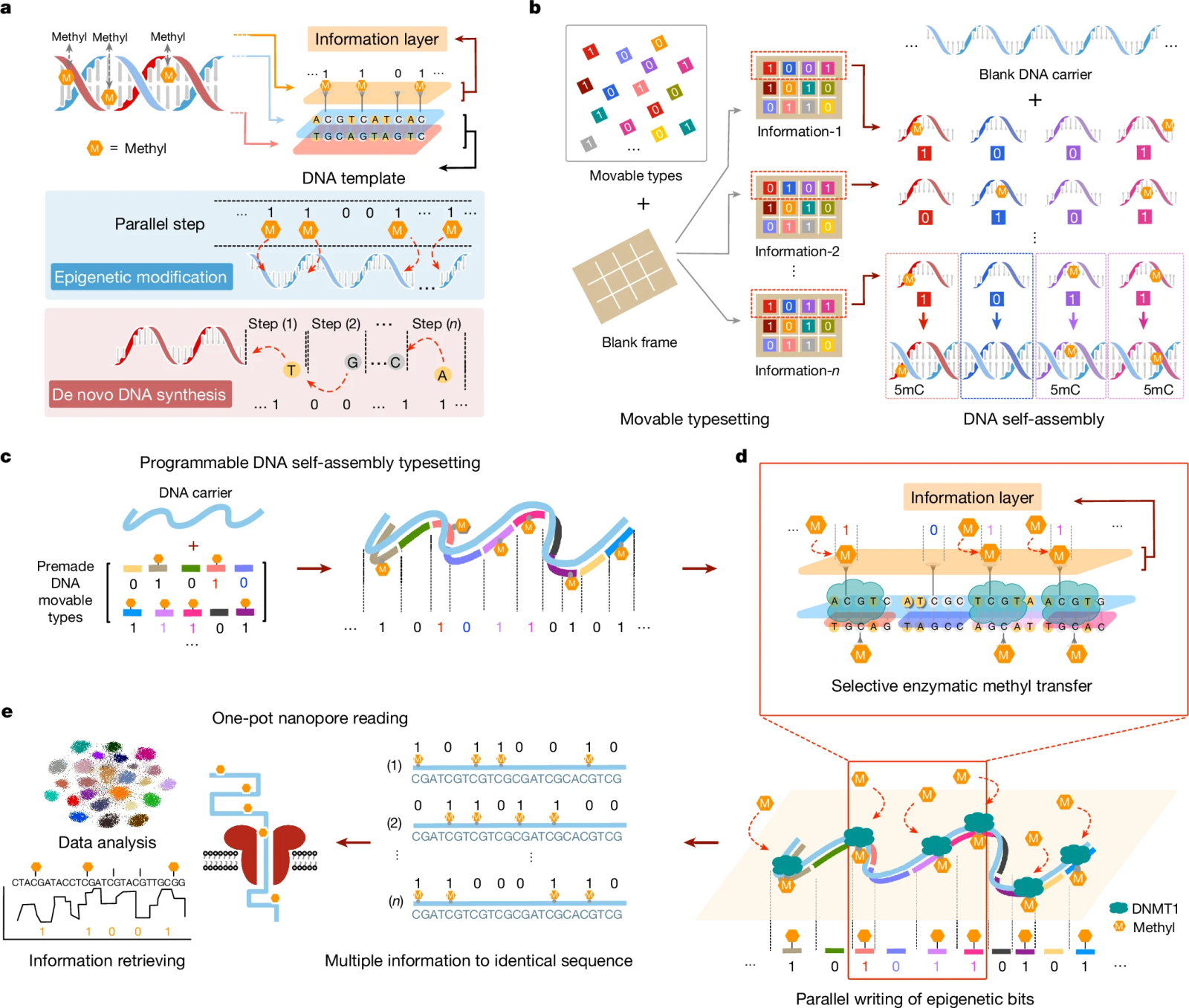
a) Mechanism of epigenetic information storage.
b) Schematic of programming mobile DNA types.
c) Programmable assembly of mobile DNA types carrying epi-bits.
d) Parallel printing via DNMT1-guided catalysis to selectively write epi-bits.
e) Nanopore sequencing of modified templates and collective methylation analysis.
This breakthrough opens promising prospects for large-scale data storage while offering a customizable and accessible solution. The work of Zhang and his team marks a significant step toward the adoption of DNA as a data storage medium, with potential applications in various fields, from archiving to bioinformatics.
What is DNA methylation?
DNA methylation is a biochemical process that involves adding a methyl group (-CH3) to a cytosine base in DNA. This epigenetic modification plays a crucial role in regulating gene expression.
In the context of data storage, methylation is used to encode information. Each methylated or non-methylated site on the DNA represents a bit of information, similar to the binary system used in computing.
This method allows data to be stored densely and stably, while remaining reversible. The enzymes responsible for methylation can be controlled to add or remove methyl groups, enabling the rewriting of stored data.
DNA methylation is also a natural process in cells, where it contributes to cell differentiation and responses to environmental stimuli. Its use for data storage is thus inspired by existing biological mechanisms.