Follow us on Google News (click on ☆)
This structure allows the migration of chromosomes during cell division, ensuring its fidelity. They showed in particular that three parameters are sufficient to explain this variability.
A data science approach to study cell variability
Each cell is different from its neighbor. Quantifying this variability is essential because it contains important information about underlying cellular mechanisms.
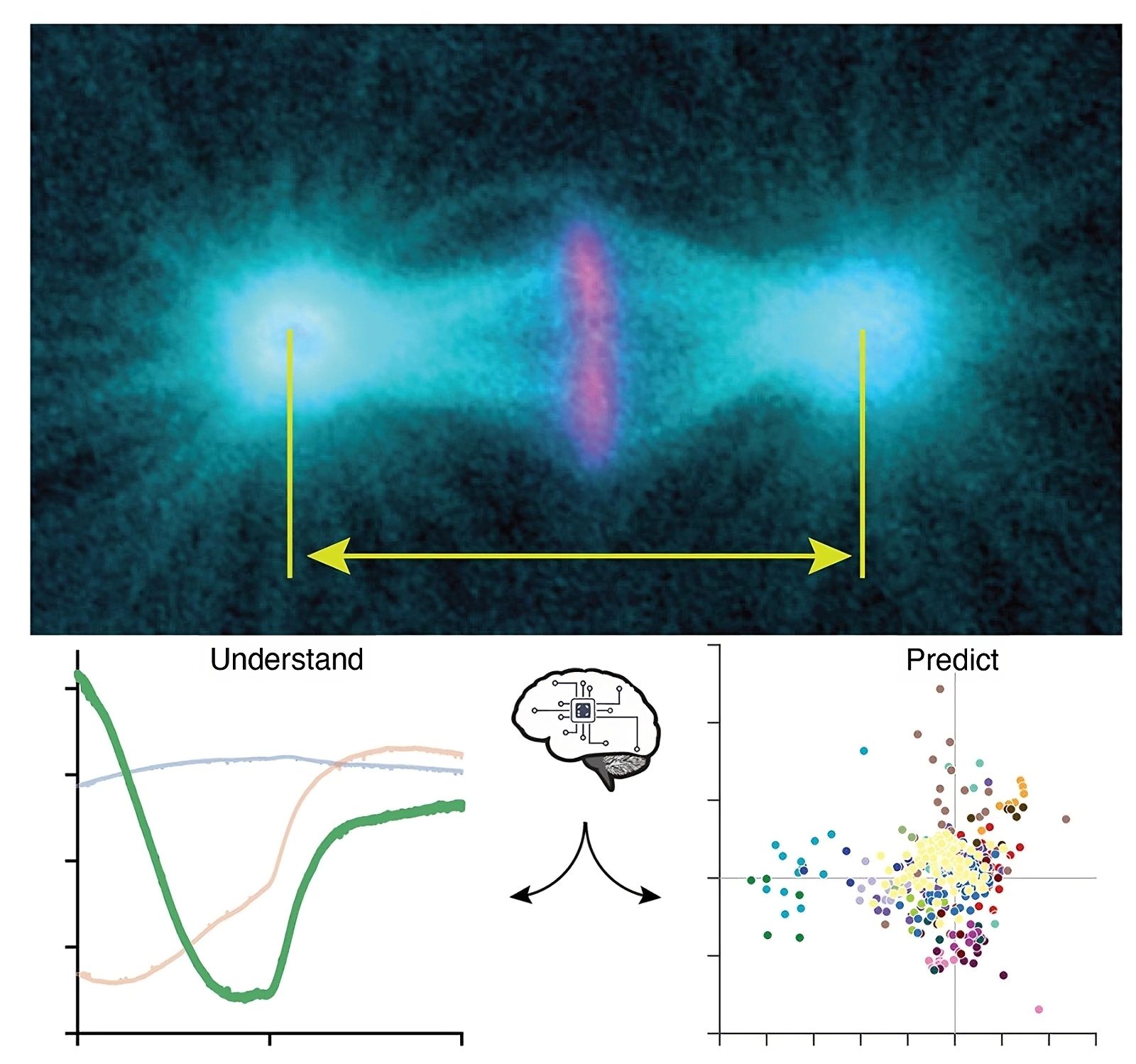
Above, microtubules marked in cyan and chromosomes in magenta, along with the mitotic spindle poles, including centrosomes, structures tracked in our analyses. The image is taken in late metaphase.
Below left, the three archetypes, with the less expected third archetype also visible. These archetypes represent typical spindle elongations over time during metaphase and anaphase.
Below right, a classification showing the dosage of each archetype to reproduce the elongations under some genetically perturbed conditions. Each color corresponds to a given gene. When points are close together, this suggests similar phenotypes, indicating that these genes may belong to the same signaling pathway or mechanism.
© Jacques Pécréaux and Laurent Chesneau
In an article published in the journal PloS Computational Biology, the scientists focused on the length of the mitotic spindle, which forms to allow the migration of chromosomes during cell division, to study this variability. This measurement is commonly used to indicate whether the division is proceeding correctly.
They used the nematode worm, Caenorhabditis elegans, to conduct the study. In this model, cell divisions are well characterized and reproducible. Additionally, its genome can be easily manipulated to precisely control the relationship between phenotype and genotype.
The scientists compiled elongation curves from 1,500 cells under control and genetically perturbed conditions to represent the variety of possibilities. To carry out an unbiased analysis, they based their approach solely on the data. The variability descriptors were automatically extracted.
With this method, they obtained two descriptors similar to those already known: spindle length and elongation rate in anaphase (the phase of mitosis where chromosomes reach the spindle poles).
However, they also discovered a new descriptor: shortening at the end of metaphase (the phase of mitosis where chromosomes are gathered in the center of the spindle) — present in all conditions. Such a phenotype had previously been limited to cells with defective chromosome attachments.
An analysis that highlights fundamental mechanisms.
These three descriptors account for 95% of the variability, suggesting that the complex choreography of the spindle relies on just a few basic mechanisms. This also limits the possible phenotypes, pointing to mechanisms ensuring the robustness of the division.
Furthermore, the scientists demonstrated that the final spindle length in anaphase, important for determining the fate of daughter cells, is already set at the end of metaphase, despite the spindle being completely rearranged between the two phases. This reveals an unexpected interdependence between metaphase and anaphase spindles.
Moreover, the same descriptors explain the variability under genetically perturbed conditions. This suggests that no new mechanisms appear in defective cells; only the contributions of existing mechanisms change.
Ultimately, these findings shed light on the fundamental mechanistic principles governing mitotic spindles and their robustness. This will help to identify the mechanisms by which cancer cells manage to divide despite accumulated defects and antimitotic treatments.
Beyond these initial results on the mitotic spindle, this work also provides a practical tool for quantifying phenotypes in other cellular contexts. Through the use of artificial intelligence, this method will suggest new candidate genes involved in cell division mechanisms. This is a significant step toward enhancing our understanding and identifying mechanism actors, to potentially develop them into future therapeutic targets.
References:
Le Cunff Y, Chesneau L, Pastezeur S, Pinson X, Soler N, Fairbrass D, et al. (2024). Unveiling inter-embryo variability in spindle length over time: Towards quantitative phenotype analysis.
PLoS Comput Biol 20(9): e1012330. https://doi.org/10.1371/journal.pcbi.1012330