Follow us on Google News (click on ☆)

Illustration image Pixabay
The stability of genetic material is essential for the proper functioning of each of our cells. However, accidents can occur, for example due to radiation or errors during DNA replication. For repairing these breaks, there exist highly conserved mechanisms across all species that are responsible for carrying out faithful repairs.
Homologous recombination is a high-fidelity repair pathway for breaks, which uses an intact DNA molecule with an identical sequence as a template. This sequence may come from the sister chromatid, the more distant homologous chromosome, or other repeated sequences within the genome. This intact homologous copy must therefore be identified within the vastness of the genome and the nuclear space. How is this search for a homologous needle in the genomic haystack conducted?
To answer this question, scientists from CNRS developed a high-throughput genomic technique called ssHi-C in yeast Saccharomyces cerevisiae, a model organism for studying fundamental DNA maintenance mechanisms. This technique allowed them to map, at the genome level, the contacts made by the break. The data revealed that the search for homology occurs in two major phases:
- a first phase in which genome structuring by cohesins constrains the search locally, with cohesins being proteins that ensure sister chromatid cohesion by trapping DNA within them.
- a second phase wherein the search frees itself from these constraints and investigates distant genomic sites. This long-range search is enabled by the production of long single-stranded DNA on both sides of the break, and their stiffening through the formation of a filament by the recombinase Rad51. This protein, which catalyzes recombination events, acts in this case like a fishing rod, casting its line made of long single-stranded DNA deep into the nucleus. The two ends of the break search in a coordinated manner, and if one end finds a homologous sequence, the other end intensifies its search in the vicinity. Surprisingly, certain regions of the genome engage with the break more frequently than others, such as chromosome III. Thus, the genome is not homogeneously queried during the homology search, suggesting the existence of targeting mechanisms that will be the subject of future studies.
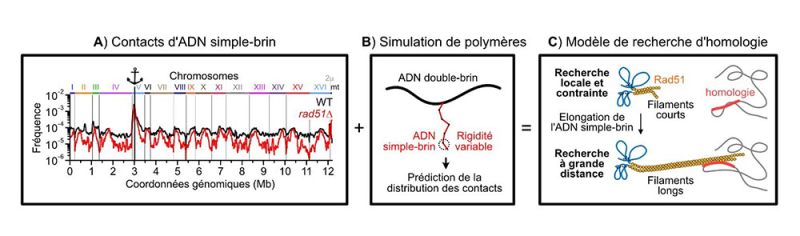
(A) The ssHi-C technique allows the determination of the contact frequency between single-stranded DNA generated at a break site (represented by the anchor) and the rest of the genome in a wild-type or mutant context.
(B) These experimental data are compared with polymer simulation predictions.
(C) This leads to a model of homology search expansion based on the formation of long rigid filaments formed by the Rad51 protein on both sides of the break.
© Aurèle Piazza
These discoveries lead to a general model of homology search based on the rigid structure of the Rad51 filament and its partners. It opens new research directions, particularly to understand the defects associated with mutations in tumor suppressor genes, such as Rad51 paralogs, in the regulation of the Rad51 filament structure and the extent of the homology search. Ultimately, the universally applicable ssHi-C technique should allow the study of the spatial localization of DNA during replication or repair in many organisms and cellular contexts.
References:
Mechanism of homology search expansion during recombinational DNA break repair in Saccharomyces cerevisiae.
A. Dumont, N. Mendiboure, J. Savocco, L. Anani, P. Moreau, A. Thierry, L. Modolo, D. Jost, A. Piazza.
Molecular Cell, August 22, 2024. DOI: 10.1016/j.molcel.2024.08.003