Follow us on Google News (click on ☆)
The search for drug candidates is primarily based on the experimental or numerical screening of as many molecules as possible that are thought to hold therapeutic virtues. For experimental screening, it's the number of molecules available for biological trials that determines the scope of screening, a scope that can be extended to other "theoretical" chemical systems through virtual screening.

The number and therapeutic performance of molecules that can successfully pass biological tests are mainly dependent on the "chemical space" screened: the larger and more chemically diverse the molecule bank is, the more effective the screening will be.
Scientists from the Therapeutic Innovation Laboratory (CNRS-University of Strasbourg) have just designed particularly efficient inhibitors of therapeutically interesting proteins by virtually screening ultra-wide theoretical chemical spaces (up to 5 billion molecules). The screening method they developed has allowed them to target for now fifteen drug candidates which they have synthesized, then biologically evaluated.
Named "SpaceDock," this innovative approach begins by identifying and locating the active sites of the protein with which the inhibitors could interact. For the dopamine D3 receptor, 135,000 commercial chemical reactants capable of anchoring to this active site were thus selected.
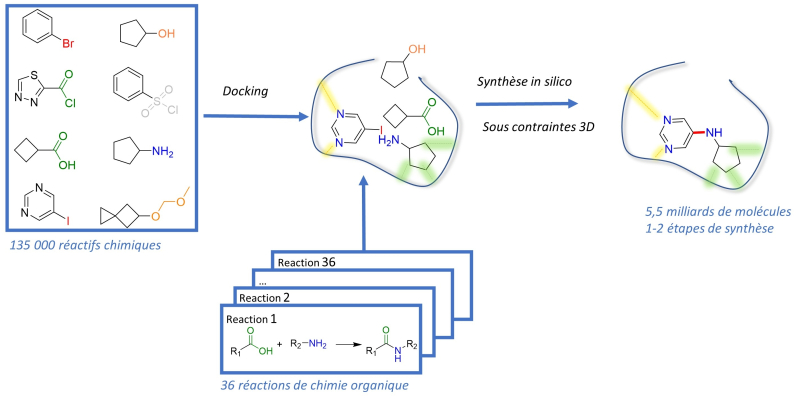
Structure of a drug candidate bound to its receptor, generated by artificial intelligence
© Didier Rognan
The scientists then took into account the topological constraints of the protein's three-dimensional structure, indicating among these reactants those that could come close enough to interact and form new molecules... Hundreds of billions of approaches are thus possible, the outcome of which can be predicted by following the procedures of 40 "classic" organic chemistry reactions. The largest screening library ever assembled!
Ultra-fast, selective and reliable, this method, published in the journal ACS Cent. Sci., allows for the prediction, synthesis, and testing of therapeutically oriented molecules by drawing from a palette of chemical substances unprecedented in its size, relevance, and diversity.
Author: CCdM
Reference:
François Sindt, Anthony Seyller, Merveille Eguida & Didier Rognan.
Protein Structure-Based Organic Chemistry-Driven Ligand Design from Ultralarge Chemical Spaces
ACS Cent. Sci. 2024
https://pubs.acs.org/doi/10.1021/acscentsci.3c01521