Follow us on Google News (click on ☆)
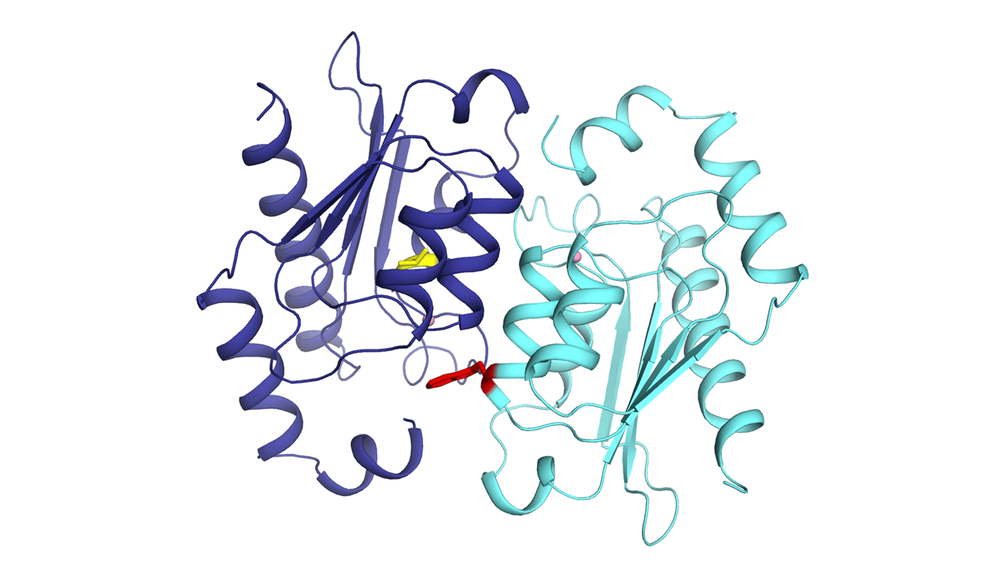
Two copies of a baker's yeast protein in a relationship of codependence. Loss-of-function mutations (in yellow and red) make them incapable of functioning without working together.
Philippe Després
For the same cellular function, the protein "machine" in humans often has more components than that in bacteria. "We might be inclined to think that these components were added by natural selection, that this added complexity serves a purpose or improves performance in humans. But that's not necessarily the case. The bacterial machine is often more efficient, even with fewer components," notes Philippe Després.
Even though complexity doesn't have an immediate negative effect on efficiency, added components do need to be produced, which requires more energy. As a result, the machine is more difficult to maintain, according to the young researcher, who conducted his thesis under the supervision of Professor Christian Landry.
A "toxic" relationship
If there is no added value, what mechanisms explain the complexity of protein machines in humans? A theoretical model from about twenty years ago suggests that random mutations might be to blame.
"At some point during evolution, the gene encoding one of the proteins in the machine may be duplicated, then undergo loss-of-function mutations. The new proteins encoded by these mutated genes would develop a codependent relationship, a toxic relationship. In other words, the two copies of the proteins must now assemble in order to perform the same function that a single ancestral copy once did," explains Philippe Després.
According to the young researcher, this theory could help explain the tendency for protein assemblies in humans to become larger over time. "This random mechanism operates somewhat against natural selection," he adds.
Proof of concept
Until now, this theoretical model of increasing complexity had never been demonstrated in the lab. To achieve this, the team used a protein from baker's yeast as a model. The researchers subjected it to mutation processes and observed that several combinations led to codependence and increased complexity.
Less than 1% of the combinations developed a codependent relationship to function—a higher percentage than the team had expected. "It seems small," admits the researcher. "But over the evolutionary timescale, this creates many opportunities for complexity-adding events. The process might be even more common than we think."
The idea of codependence in protein machines could have significant implications for cancer research. "In cancer cells, the two copies can mutate independently. Previously, if both copies lost their function, we might have assumed the gene was no longer functional. Now we have to determine if these copies can still assemble to form a functional machine," explains Philippe Després, now a postdoctoral researcher at the Broad Institute in Boston and the University of Guelph, Ontario.
The "cherry on top," according to the researcher, was analyzing the molecular structure of the proteins down to the atom. "This is an unprecedented resolution to study how the two proteins assemble to function despite the loss-of-function mutations." He pointed out the collaboration of Professor Rong Shi's team from the Department of Biochemistry, Microbiology, and Bioinformatics at Université Laval.
The other co-authors of the study published in Science include Alexandre K. Dubé, Marie-Ève Picard, Jordan Grenier, Rong Shi, and Christian Landry.